Groundwater model
groundwater-model.Rmd
library(nitratesgame)
library(dplyr)
library(tidyr)
library(sf) # needed for package to represent well capture polygons
library(units) # needed for package, ensures proper units
library(ggforce) # for plotting axes with units
library(patchwork) # for aligning axes of multiple subplotsGroundwater model
First we set some basic parameters for the scenario including:
- Household water use
- Seepage rate
- Annual precipitation
- Seepage fraction
- Seepage fraction
- Well source area
- radius of source area
- Housing density
# Get area of hh withdrawal
# hh_annual <- set_units(76, "gallon/day") %>% # from USGS water data
# set_units("ft^3/year") * 4
# precip_annual <- set_units(1070, "mm / year") #%>%
# seepage_fraction <- 0.4
# seepage_annual <- precip_annual * seepage_fraction
# area_of_withdrawal <- hh_annual / seepage_annual
# wells are screened from 10 ft to 20 ft depth below the water table
z1 <- set_units(10,"ft")
z2 <- set_units(20,"ft")
rs <- set_units(20, "ft") # this is the radius of the region of well capture
density <- set_units(0.5, "1/acre") # the housing densityNow we need a grid of septic systems. You can create an array of
wells yourself or use get_hh_grid. The inputs to
get_hh_grid are density and area
as units objects.
area <- set_units(64, "acre")
hh_array <- get_hh_grid(density, area)
hh_array$id <- 1:nrow(hh_array) # supply an idThe groundwater model predicts the probability that a point source
will contaminate at least one well in an array of wells. However, in
this case we are interested in the probability that an array of septic
fields (considered point sources) will lead to contamination in a single
well. This problem can be modeled using
get_intersection_probability by considering the well as a
point source and the septic systems as “virtual wells” such that the
spatial relationship between a well and septic system (point source) is
the same as the relationship between the virtual well and well (now
considered the point source). The function
get_septic_well_array does this job for us. See the
?get_septic_well_array for details.
virtual_well_array <- get_septic_well_array(hh_array, "septic", z_range = c(z1, z2), rs = rs)Here you can see that the virtual well array is identical to the septic array but rotated by 180 degrees around the origin.
library(ggplot2)
library(ggforce) # needed to plot axes using units objects
ggplot(mapping = aes(x, y, color = id)) +
geom_point(data = hh_array, aes(shape = "septic systems"), size = 4, stroke = 1) +
geom_point(data = virtual_well_array, aes(shape = "virtual wells"), size = 2) +
scale_shape_manual(values = c(1, 16)) +
scale_color_viridis_c(option = "B") + coord_equal()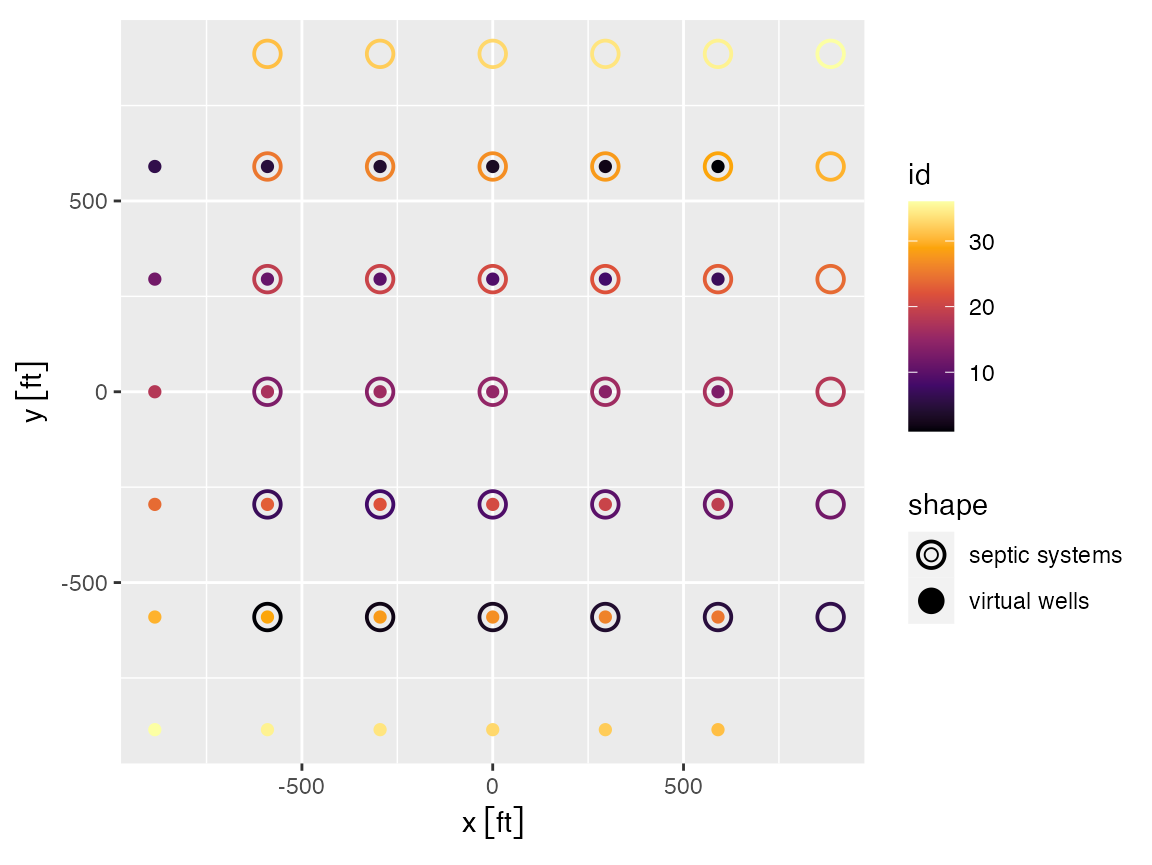
The array virtual_well_array contains sf geometries for
each of the wells in \(\theta-\phi\)
space. We can plot these geometries projected onto the z-axis such that
it appears as viewed from the point source at the land surface. To do so
we can take \(z_{projected}=\sin(\phi)\) and plot using
geom_rect from ggplot. We zoom in only on
\(\theta \in [0,pi/2]\).
p_wells <- ggplot(virtual_well_array) +
geom_rect(aes(xmin = theta1, xmax = theta2, ymin = sin(phi1), ymax = sin(phi2), fill = id), color = NA, alpha = 1) +
scale_fill_viridis_c("id") +
scale_y_reverse("sin(phi)") +
scale_x_continuous(breaks = c(0, pi/2), labels = c("0", "pi/2")) +
coord_cartesian(xlim = c(0, pi/2))
p_phi <- ggplot(data.frame(alpha = seq(0,20,by=.1)) %>% dplyr::mutate(phi = atan(1/alpha))) +
stat_density(aes(sin(phi))) +
scale_x_reverse("sin(phi)") +
coord_flip()
p_wells + p_phi + patchwork::plot_layout(widths=c(0.8,0.2))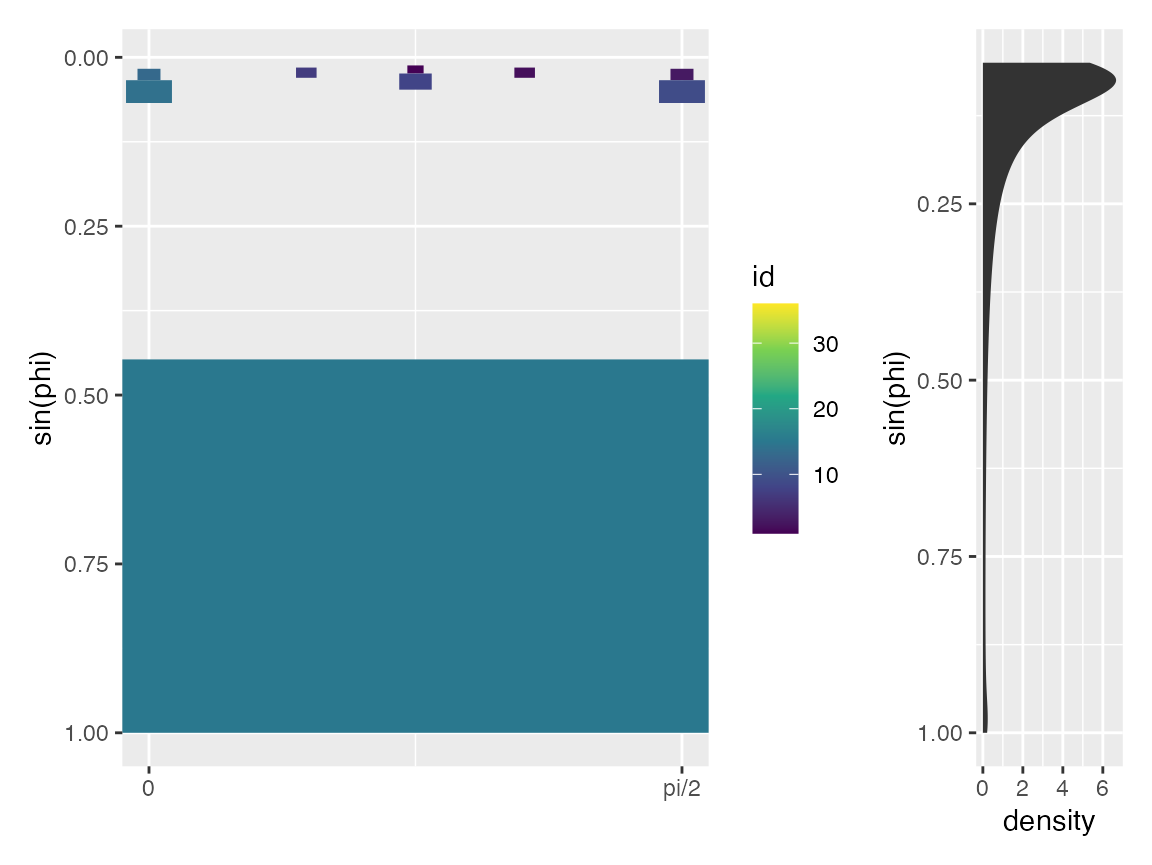
Get probability
Now get the probability of contamination of the well, using the virtual well array.
gw_example <- virtual_well_array %>%
get_intersection_probability(theta_range = c(0,pi/2), alpha_range = c(0, 20),
self_treat = FALSE, show_progress = FALSE)
gw_example
#> [1] 0.1225776Get multiple probabilities at once
Now, let’s get probabilities for each row of a tibble
which contains columns for all of the appropriate input variables. Note
that this must be a tibble and not a
data.frame, because it has to store complex objects (i.e.,
list columns).
# Create a single row with constant parameters
params_row <- tibble(
z1 = set_units(10, "ft"),
z2 = set_units(20, "ft"),
area = set_units(64, "acre"),
theta_range = list(c(0, pi/4)), # this will be unlisted in the function
alpha_range = list(c(0, 20)), # this will be unlisted in the function
hh_array_type = "septic")
# Add varying parameters
params_df <- params_row %>%
crossing(density = set_units(c(0, 0.5), "1/acre"),
rs = set_units(c(10, 20),"ft"),
self_treat = c(TRUE, FALSE)) # if self_treat is TRUE, a household cannot contaminate its own well
# Get probabilities
params_df$probs <- get_contamination_probabilities(params_df)
params_df[,c("density", "rs", "self_treat", "probs")]
#> # A tibble: 8 × 4
#> density rs self_treat probs
#> [1/acre] [ft] <lgl> <dbl>
#> 1 0 10 FALSE 0.05
#> 2 0 10 TRUE 0
#> 3 0 20 FALSE 0.1
#> 4 0 20 TRUE 0
#> 5 0.5 10 FALSE 0.0613
#> 6 0.5 10 TRUE 0.0113
#> 7 0.5 20 FALSE 0.123
#> 8 0.5 20 TRUE 0.0226